고려대학교 교수소개
Knowledge & Innovation
소개

Prof. 우재성
Tel: 02-3290-3406
E-mail: jaesungwoo@korea.ac.kr
- About Professor
- Curriculum Vitae
- Publication
- Research
Profile
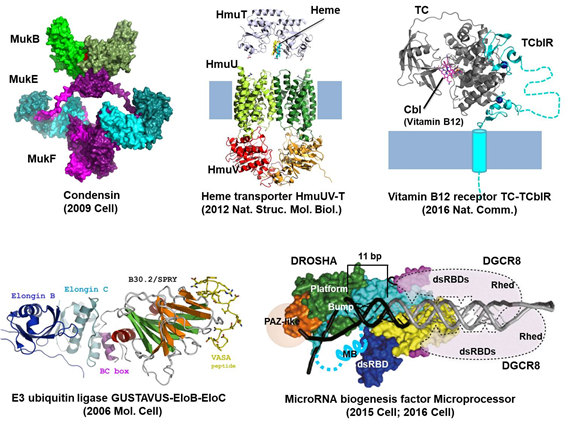
Structural Biology of Molecular Machines.
Proteins in the cell function like machines and buildings in our daily life. For example, an actin filament is a road for movement of proteins or vesicles, and myosins walk on the road. Condensin organizes and condenses chromatin threads like a tailor sews a chain stitch. Drosha and Dicer, like cutter machines, measure the length of and cleave double-stranded RNA. To fully understand how proteins work, their structural informations are absolutely required. It would be great if we could directly see intact proteins through a microscope, but the current technology is far from viewing them in high resolution. Instead, we can solve the average structure of purified proteins by high-end technologies such as X-ray crystallography or cryo-electron microscopy (cryo-EM). We use these techniques to solve high resolution structures of dynamic protein complexes with a focus on cell junction proteins and RNA-binding proteins.
Curriculum Vitae
Education
- 2003-2007 Ph.D., POSTECH, Pohang, Republic of Korea
- 1996-2002 B.S., POSTECH, Pohang, Republic of Korea
Professional Experiences
- 2017-present Associate professor, Korea University, Republic of Korea
- 2013-2017 Research Professor, Seoul National University, Republic of Korea
- 2013-2017 Research Fellow, Institute for Basic Science, Republic of Korea
- 2009-2013 Postdoctoral fellow, ETH Zurich, Zurich, Switzerland
- 2007-2009 Postdoctoral fellow, POSTECH, Pohang, Republic of Korea
Publication
- Alam A, Woo J-S, Schmitz J, Prinz B, Root K, Chen F, Bloch JS, Zenobi R, Locher KP (2016)
- Structural basis of transcobalamin recognition by human CD320 receptor
- Nature Communications 7(May):12100
- Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng S, Kim VN, Woo JS (2016)
- Structure of Human DROSHA
- Cell 164(1–2):81–90
- Lee J, Park I-S, Kim H, Woo J-S, Choi B-S, Min D-H (2015)
- BSA as additive: A simple strategy for practical applications of PNA in bioanalysis
- Biosensors and Bioelectronics 69:167–173
- Nguyen TA, Jo MH, Choi Y-G, Park J, Kwon SC, Hohng S, Kim VN, Woo J-S (2015)
- Functional Anatomy of the Human Microprocessor
- Cell 161(6):1374–1387
- Nguyen TA, Kwon SC, Woo J (2015)
- How human Microprocessor recognizes primary microRNAs and finds the correct cleavage sites
- Bio Design 77–87
- Woo J-S, Kim VN (2014)
- MeCP2 caught moonlighting as a suppressor of MicroRNA processing.
- Developmental Cell 28(5):477–8
- Cha HJ, Jang DS, Kim YG, Hong BH, Woo JS, Kim KT, Choi KY (2013)
- Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
- Molecules and Cells 36(1):39–46
- Suh HY, Kim JH, Woo JS, Ku B, Shin EJ, Yun Y, Oh BH (2012)
- Crystal structure of DeSI-1, a novel deSUMOylase belonging to a putative isopeptidase superfamily
- Proteins: Structure, Function and Bioinformatics 80(8):2099–2104
- Woo J-S, Zeltina A, Goetz B a, Locher KP (2012)
- X-ray structure of the Yersinia pestis heme transporter HmuUV.
- Nature Structural & Molecular Biology 19(12):1310–5
- Kugler J-M, Woo J-S, Oh B-H, Lasko P (2010)
- Regulation of Drosophila vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors.
- Molecular and Cellular Biology 30(7):1769–1782
- Mattle D, Zeltina A, Woo JS, Goetz BA, Locher KP (2010)
- Two Stacked Heme Molecules in the Binding Pocket of the Periplasmic Heme-Binding Protein HmuT from Yersinia pestis
- Journal of Molecular Biology 404(2):220–231
- Suh H-Y, Lee D-W, Lee K-H, Ku B, Choi S-J, Woo J-S, Kim Y-G, Oh B-H (2010)
- Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA.
- The EMBO Journal 29(2):496–504
- Shin HC, Lim JH, Woo JS, Oh BH (2009)
- Focal localization of MukBEF condensin on the chromosome requires the flexible linker region of MukF
- FEBS Journal 276(18):5101–5110
- Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, Ha NC, Oh BH (2009)
- Structural Studies of a Bacterial Condensin Complex Reveal ATP-Dependent Disruption of Intersubunit Interactions
- Cell 136(1):85–96
- Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH (2008)
- Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine γ-herpesvirus 68
- PLoS Pathogens 4(2)
- Woo J-S, Imm J-H, Min C-K, Kim K-J, Cha S-S, Oh B-H (2006)
- Structural and functional insights into the B30.2/SPRY domain.
- The EMBO Journal 25(6):1353–1363
- Woo JS, Suh HY, Park SY, Oh BH (2006)
- Structural Basis for Protein Recognition by B30.2/SPRY Domains
- Molecular Cell 24(6):967–976
- Woo J-S, Jung J-S, Ha N-C, Shin J, Kim K-H, Lee W, Oh B-H (2003)
- Unique structural features of a BCL-2 family protein CED-9 and biophysical characterization of CED-9/EGL-1 interactions.
- Cell Death and Differentiation 10(12):1310–9
Research
Research Topic
Structures and permeation mechanisms of cell junction proteins
Maintenance of multicellularity in animals basically requires intercellular attachment, metabolite transport and communication, which are facilitated by cell junctions. While anchoring junctions and occluding junctions are responsible for strong adherence and barrier formation between adjacent cells, gap junctions mediate the direct intercellular passage of small molecules such as ions, metabolites, and signal molecules. Molecular transport through gap junctions is crucial for many cellular and physiological processes including development, cell death, and electrical signal transmission in the brain and heart. A gap junction channel (GJC) comprises 12 connexin proteins. In human cells, 21 different connexins form structurally and functionally diverse GJCs through their homomeric and heteromeric interactions. Interestingly, the permeability of GJC is tightly regulated by many factors such as voltage, pH, divalent ions, and posttranslational modifications. However, the molecular mechanisms underlying hetero-oligomerization and permeability regulation are mostly unknown. Our structural study on human GJCs aims at visualizing the interaction among three cytosolic domains of connexin as well as their interaction with the channel pore. The structure-based biochemical study will reveal the detailed molecular mechanism of permeability regulation. We will also investigate the rule of connexin assembly to understand the heterogeneous nature of human GJCs.
Structures and RNA-processing mechanisms of microRNA biogenesis factors
MicroRNAs (miRNAs) inhibit gene expression through the interaction with their target mRNAs. For the last decade, more than 1000 miRNAs have been found to be encoded in a human cell and thus regulate virtually all cellular processes. Despite the general importance of miRNAs in biology and medical science, the molecular mechanism of miRNA biogenesis is not fully understood. Our study focuses on the primary miRNA (pri-miRNA) processing step mediated by the Drosha-DGCR8 (Microprocessor) complex, in which the miRNA sequence and its target mRNAs are determined. The ultimate goal is to completely understand how Microprocessor determines the cleavage sites for diverse pri-miRNAs.
Development of the human cell expression system for structural biology
Majority of human proteins, especially membrane proteins and secreted proteins, are not functionally expressed in the conventional expression systems such as E. coli, yeast, and insect cell systems. To produce and purify human proteins in the milligram scale, we have developed human cell expression systems. The current system has been successfully used to produce Microprocessor and several membrane transporters and channels. We are further developing this system for mass production of large secreted proteins.
